OMICs technologies uncovered several unknown aspects of central dogma
Several scientific and technological advances uncovered new knowledge about each step in central dogma – unidirectional flow of information from DNA to RNA to Proteins and in many cases from RNA to Proteins. Simultaneously, these advances discovered epigenetic regulation of central dogma and the importance of probing other biomolecules such as lipids and metabolites.
Due to rapidly evolving technologies and the reducing cost of generating OMICs data – quantitative high throughput data on biomolecules, we require specific tools to analyze it rapidly. Data analysis is a key factor in R&D processes due to the increasing resolution of these measurements in spatio-temporal dimensions from organism to tissues and even to individual cells. This is also evident from the large investments OMICs data attracts across the R&D industry and academia.1 In Jan 2023, EU launched a joint program worth 16.5 million euros for large scale analysis of OMICs data for drug-target finding in neurodegenerative diseases alone.
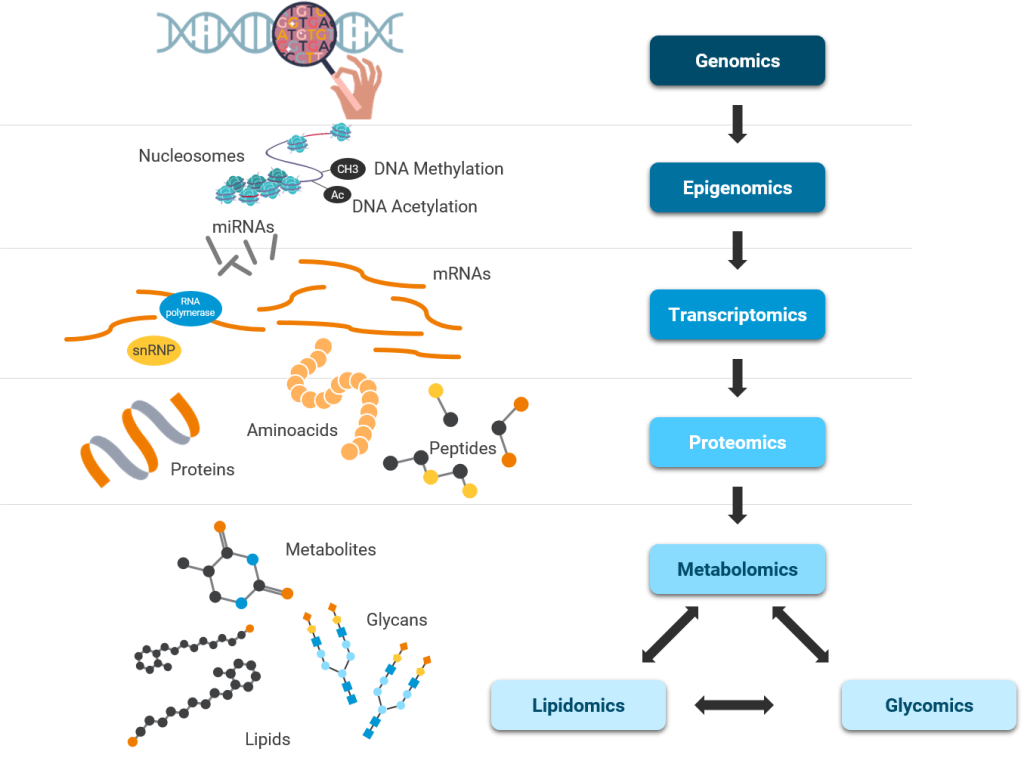
Reproducibility and Ease of Operations are major challenges for OMICs Data Analysis
Scientific workflows or pipelines (Figure 2) – series of software tools working in a stepwise manner one after the other – are important to rapidly analyze and interpret vast amounts of data generated using various OMICs techniques. For example, RNA-seq data analysis (Transcriptomics) involves trimming, aligning, quantification, normalization and differential gene expression analysis where output of one tool serves as input for the next tool in the workflow. Permutations of these tools can lead to over 150 sequential workflows or pipelines, so reproducing and comparing their results can be challenging2.
Below are some examples of pipelines and software tools to perform different analysis steps in different OMICs experiments.
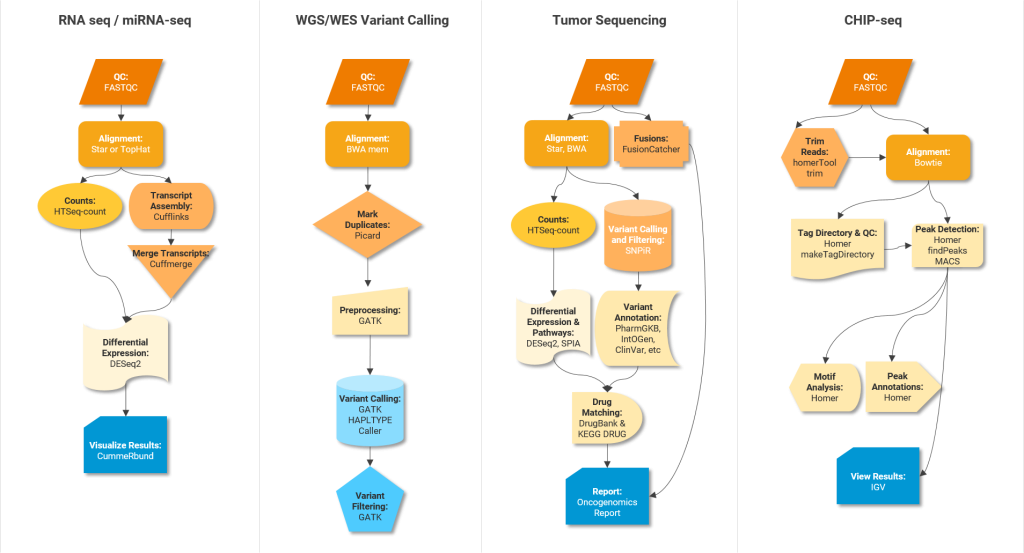
Integrative frameworks can help facilitate the execution of these pipelines. Typical no/low code frameworks for non-programmers are Galaxy, Unipro UGENE NGS and MIGNON. For developers, suitable analysis frameworks include Snakemake, Nextflow and Bpipe. Community driven platforms like nf-core provide peer-reviewed best practice analysis pipelines written in Nextflow.
One way to ensure efficient/streamlined R&D is to establish and follow standard practices for OMICs data analysis. Figure 2 shows gold standard software tools for performing various intermediate steps in different OMICs data analysis pipelines. Standardized OMICs practices – tools and frameworks – will facilitate accessibility (A in FAIR) and reproducibility of high quality results. It will also enhance their business value (please also refer to the following blog post: Multi-Omics Data Integration in Drug Discovery). We would like to point to the analysis pipelines for major OMICs assay types developed by the ENCODE Data Coordinating Center (DCC).
Although uniformity is valuable in OMICs data analyses, customization is also equally valuable for specific scientific contexts. Different OMICs experiments require different handling of the data and analyses. For example, high variation in signal-to-noise ratio in ChIP-seq experiments to identify transcription factor (TF) binding sites necessitates a wide range of quality thresholds. RNA-seq data analyses are driven by factors such as read size, polyadenylation status, strandedness and require different parameters or settings. Hence, there are multiple “generic” factors that can be standardized, while individual parameters and settings can be customized to suit specific scientific questions.
Factors to consider while standardizing OMICs data analysis workflows:
- Knowledge and understanding of central dogma and design of various high throughput experiments
- Standard data files and formats (e.g. using common reference genome across labs/departments where ever possible)
- Domain specific language such as Workflow Description Language (WDL) and/or Common Workflow Language (CWL) to enhance Interoperability (I of the FAIR principles)
- Flexible frameworks that can run locally (e.g. on HPC) and in cloud (e.g. AWS, Azure, Google)
- Common framework for testing the workflows
- Wrappers for automated file handling (import and export of data, parameters)
- User friendly interfaces for interactive usage
- Platform-agnostic installation of frameworks, packages, libraries
- Common package managers like Conda
- Intuitive and interactive visualization of workflows, their progress and their results
- Metadata tracking
- Common and portable sharing mechanism for pipelines (e.g. Docker images), data and results
Uniform OMICs Data Analysis Workflows to Empower your Future:
Relatively well-established OMICs data generation techniques demand standardized ways of data analysis while allowing customization necessary for a specific scientific context. To accelerate discovery and actionable data-driven decisions within your organization, take a step closer to FAIR OMICs data by establishing gold standard workflows and frameworks for OMICs data analyses.
How Can OSTHUS Help?
- Scientific advise (in experimental design)
- Tool selection for your specific scientific question
- Developing an automated workflow using different workflow management frameworks
- Customizing an existing workflow by developing scripts for individual steps
- Visualizing analysis results using available business intelligence tools and/or developing bespoke interfaces suitable to answer specific questions
Contact us to know more about how we are instrumental in our customers’ goal
to derive scientific insights from their OMICs data.
References:
- https://biotechfinance.org/q2/
- Corchete LA et al, Scientific Reports, 2020
Disclaimer
The contents of this blog are solely the opinion of the author and do not represent the opinions of PharmaLex GmbH or its parent Cencora Inc. PharmaLex and Cencora strongly encourage readers to review the references provided with this blog and all available information related to the topics mentioned herein and to rely on their own experience and expertise in making decisions related thereto.





